Developing a toolkit to study branched ubiquitin chains!
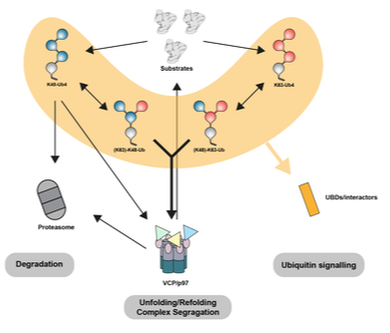
Lange SM, McFarland MR, Lamoliatte F, Kwaśna D, Shen L, Wallace I, Cole I, Armstrong LA, Knebel A, Johnson C, De Cesare V & Kulathu Y. (2023) "Comprehensive approach to study branched ubiquitin chains reveals roles for K48-K63 branches in VCP/p97-related processes"
bioRxiv
Branched ubiquitin (Ub) chains make up a significant proportion of Ub polymers in human cells and are formed when two or more sites on a single Ub molecule are modified with Ub creating bifurcated architectures. Despite their abundance, we have a poor understanding of the cellular functions of branched Ub signals that stems from a lack of facile tools and methods to study them. In this study, we develop a comprehensive pipeline to define branched Ub function. First, we investigate how branched chains are decoded in cells and idenitfy branched chain specific binding proteins. To study how DUBs process branched chains, we develop a quantitative assay that monitors the linkages within polyUb that are cleaved and discover debranching enzymes. Finally, we engineer nanobodies that bind to branched chains with high affinity and specificity. Applying these methods and tools we reveal roles for K48-K63 branched chains in VCP/p97-related processes. We provide a blueprint to investigate branched Ub function that can be readily applied to study other branched chain types.
Link to preprint
bioRxiv
Branched ubiquitin (Ub) chains make up a significant proportion of Ub polymers in human cells and are formed when two or more sites on a single Ub molecule are modified with Ub creating bifurcated architectures. Despite their abundance, we have a poor understanding of the cellular functions of branched Ub signals that stems from a lack of facile tools and methods to study them. In this study, we develop a comprehensive pipeline to define branched Ub function. First, we investigate how branched chains are decoded in cells and idenitfy branched chain specific binding proteins. To study how DUBs process branched chains, we develop a quantitative assay that monitors the linkages within polyUb that are cleaved and discover debranching enzymes. Finally, we engineer nanobodies that bind to branched chains with high affinity and specificity. Applying these methods and tools we reveal roles for K48-K63 branched chains in VCP/p97-related processes. We provide a blueprint to investigate branched Ub function that can be readily applied to study other branched chain types.
Link to preprint
A Guide to UFMylation
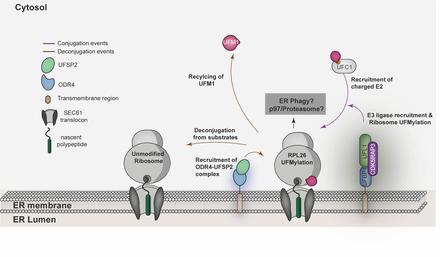
Millrine D, Peter JJ & Kulathu Y. (2023) "A Guide to UFMylation: an emerging posttranslational modification"
FEBS Journal
Ubiquitin Fold Modifier-1 (UFM1) is a ubiquitin-like modifier (UBL) that is posttranslationally attached to lysine residues on substrates via a dedicated system of enzymes conserved in most eukaryotes. Despite the structural similarity between UFM1 and ubiquitin, the UFMylation machinery employs unique mechanisms that ensure fidelity. While physiological triggers and consequences of UFMylation are not entirely clear, its biological importance is epitomized by mutations in the UFMylation pathway in human pathophysiology including musculoskeletal and neurodevelopmental diseases. Some of these diseases can be explained by the increased endoplasmic reticulum (ER) stress and disrupted translational homeostasis observed upon loss of UFMylation. The roles of UFM1 in these processes likely stem from its function at the ER where ribosomes are UFMylated in response to translational stalling. In addition, UFMylation has been implicated in other cellular processes including DNA damage response and telomere maintenance. Hence, the study of UFM1 pathway mechanics and its biological function will reveal insights into fundamental cell biology and is likely to afford new therapeutic opportunities for the benefit of human health. In this review, we provide a comprehensive guide to the current state of knowledge of UFM1 biogenesis, conjugation, and function with an emphasis on the underlying mechanisms..
Link to review
FEBS Journal
Ubiquitin Fold Modifier-1 (UFM1) is a ubiquitin-like modifier (UBL) that is posttranslationally attached to lysine residues on substrates via a dedicated system of enzymes conserved in most eukaryotes. Despite the structural similarity between UFM1 and ubiquitin, the UFMylation machinery employs unique mechanisms that ensure fidelity. While physiological triggers and consequences of UFMylation are not entirely clear, its biological importance is epitomized by mutations in the UFMylation pathway in human pathophysiology including musculoskeletal and neurodevelopmental diseases. Some of these diseases can be explained by the increased endoplasmic reticulum (ER) stress and disrupted translational homeostasis observed upon loss of UFMylation. The roles of UFM1 in these processes likely stem from its function at the ER where ribosomes are UFMylated in response to translational stalling. In addition, UFMylation has been implicated in other cellular processes including DNA damage response and telomere maintenance. Hence, the study of UFM1 pathway mechanics and its biological function will reveal insights into fundamental cell biology and is likely to afford new therapeutic opportunities for the benefit of human health. In this review, we provide a comprehensive guide to the current state of knowledge of UFM1 biogenesis, conjugation, and function with an emphasis on the underlying mechanisms..
Link to review
How is UFMylation activated and regulated?
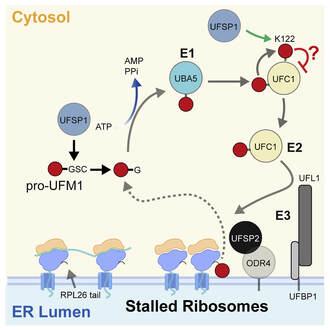
Millrine D, Cummings T, Matthews SP, Peter JJ, Magnussen H, Macartney T, Lamoliatte F, Knebel A & Kulathu Y. (2022) "Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation"
Cell Reports
An essential first step in the post-translational modification of proteins with UFM1, UFMylation, is the proteolytic cleavage of pro-UFM1 to expose a C-terminal glycine. Of the two UFM1-specific proteases (UFSPs) identified in humans, only UFSP2 is reported to be active, since the annotated sequence of UFSP1 lacks critical catalytic residues. Nonetheless, efficient UFM1 maturation occurs in cells lacking UFSP2, suggesting the presence of another active protease. We herein identify UFSP1 translated from a non-canonical start site to be this protease. Cells lacking both UFSPs show complete loss of UFMylation resulting from an absence of mature UFM1. While UFSP2, but not UFSP1, removes UFM1 from the ribosomal subunit RPL26, UFSP1 acts earlier in the pathway to mature UFM1 and cleave a potential autoinhibitory modification on UFC1, thereby controlling activation of UFMylation. In summary, our studies reveal important distinctions in substrate specificity and localization-dependent functions for the two proteases in regulating UFMylation.
Full Text
Previously on Biorxiv Now published in Cell Reports
Cell Reports
An essential first step in the post-translational modification of proteins with UFM1, UFMylation, is the proteolytic cleavage of pro-UFM1 to expose a C-terminal glycine. Of the two UFM1-specific proteases (UFSPs) identified in humans, only UFSP2 is reported to be active, since the annotated sequence of UFSP1 lacks critical catalytic residues. Nonetheless, efficient UFM1 maturation occurs in cells lacking UFSP2, suggesting the presence of another active protease. We herein identify UFSP1 translated from a non-canonical start site to be this protease. Cells lacking both UFSPs show complete loss of UFMylation resulting from an absence of mature UFM1. While UFSP2, but not UFSP1, removes UFM1 from the ribosomal subunit RPL26, UFSP1 acts earlier in the pathway to mature UFM1 and cleave a potential autoinhibitory modification on UFC1, thereby controlling activation of UFMylation. In summary, our studies reveal important distinctions in substrate specificity and localization-dependent functions for the two proteases in regulating UFMylation.
Full Text
Previously on Biorxiv Now published in Cell Reports
Chemical biology tools to study Deubiquitinases and Ubl proteases
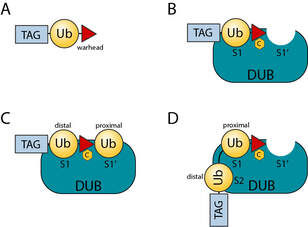
Gorka M, Magnussen H and Kulathu Y. (2022)
Seminars in Cell and Developmental Biology
The reversible attachment of ubiquitin (Ub) and ubiquitin like modifiers (Ubls) to proteins are crucial post-translational modifications (PTMs) for many cellular processes. Not only do cells possess hundreds of ligases to mediate substrate specific modification with Ub and Ubls, but they also have a repertoire of more than 100 dedicated enzymes for the specific removal of ubiquitin (Deubiquitinases or DUBs) and Ubl modifications (Ubl-specific proteases or ULPs). In this review we discuss how chemical biology has led to the development of activity-based probes and substrates that have been invaluable to the study of DUBs and ULPs. We summarise our currently available toolbox, highlight the main achievements and give an outlook of how these tools may be applied to gain a better understanding of the regulatory mechanisms of DUBs and ULPs.
Full Text
Seminars in Cell and Developmental Biology
The reversible attachment of ubiquitin (Ub) and ubiquitin like modifiers (Ubls) to proteins are crucial post-translational modifications (PTMs) for many cellular processes. Not only do cells possess hundreds of ligases to mediate substrate specific modification with Ub and Ubls, but they also have a repertoire of more than 100 dedicated enzymes for the specific removal of ubiquitin (Deubiquitinases or DUBs) and Ubl modifications (Ubl-specific proteases or ULPs). In this review we discuss how chemical biology has led to the development of activity-based probes and substrates that have been invaluable to the study of DUBs and ULPs. We summarise our currently available toolbox, highlight the main achievements and give an outlook of how these tools may be applied to gain a better understanding of the regulatory mechanisms of DUBs and ULPs.
Full Text
Mechanistic insights into protein UFMylation
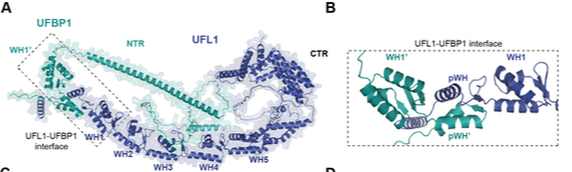
Peter JJ, Magnussen HM, DaRosa PA, Millrine D, Matthews SP, Lamoliatte F, Sundaramoorthy R, Kopito RR and Kulathu Y. (2022) "Non canonical scaffold-type ligase complex mediates protein UFMylation" Biorxiv
Protein UFMylation is emerging as a posttranslational modification essential for endoplasmic reticulum and cellular homeostasis. Despite its biological importance, we have a poor understanding of how UFM1 is conjugated onto substrates. Here, we use a rebuilding approach to define the minimal requirements of protein UFMylation. We find that the reported E3 ligase UFL1 is inactive on its own and identify UFBP1 to bind UFL1 to form an active E3 ligase complex. While UFC1 is an intrinsically Cys-reactive E2, we do not identify any catalytic cysteines on UFL1/UFBP1, suggesting a scaffold-type E3 ligase mechanism. Interestingly, the E3 ligase complex consists of winged-helix (WH) domain repeats that activate UFC1 for aminolysis. We identify the adaptor protein CDK5RAP3 to bind to and regulate E3 ligase activity potentially by preventing off-target UFMylation. In summary, our work identifies the minimal requirements for UFMylation and reveals regulatory principles of this atypical E3 ligase complex.
Link to preprint
Now published in EMBO journal
Position available! We are looking for postdocs to work on mechanistic aspects of how UFMylation maintains ER homeostasis. Please get in touch if you are interested.
Mechanisms of Deubiquitinases
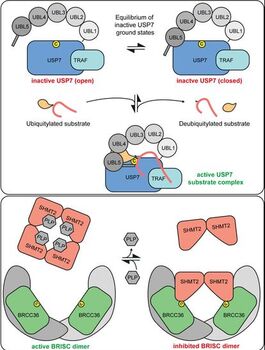
Lange SM, Armstrong L, Kulathu Y. (2021) “Deubiquitinases: from mechanisms to their inhibition by small molecules” Molecular Cell
In this comprehensive review we review the mechanisms of DUBs and how they regulate ubiquitin signalling. We also provide an overview of inhibitors to DUBs and propose a standardized nomenclature classifying inhibitors by their mode of action.
Link to review
Structure of the Pseudokinase IRAK3 suggests an allosteric mechanism for negative regulation
|
Lange SM, Nelen MI, Cohen P, Kulathu Y. (2020) Structure.
Interleukin-1 receptor associated kinases (IRAKs) are key players in innate immune signaling that mediate the host response to pathogens. In contrast to the active kinases IRAK1 and IRAK4, IRAK2 and IRAK3 are pseudokinases lacking catalytic activity and their functions are poorly understood. IRAK3 is thought to be a negative regulator of innate immune signaling and mutations in IRAK3 are associated with asthma and cancer. This paper reveals the structure of the pseudokinase IRAK3 and suggests a model for how this dead kinase may modulate IRAK kinase activity in innate immune signalling Full Text |
Catalytic mechanism of the MINDY family DUBs
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Grawert TW, Knebel A, Svergun DI and Kulathu Y. (2021) “Mechanism of activation and regulation of Deubiquitinase activity in MINDY1 and MINDY2”. Molecular Cell
MINDY1 and 2 recognize and cleave and long K48-linked polyubiquitin chains. In this work, we make the surprising finding that MINDY1/2 have 5 distinct ubiquitin binding sites within their catalytic domain that they use to trim K48 chains. We also reveal that MINDY1/2 employ an unusual catalytic mechanism and how their catalytic activity is modulated.
Full Text
Preprint
Characterizing Nsp3 from SARS CoV2 and engineering nanobodies that inhibit it
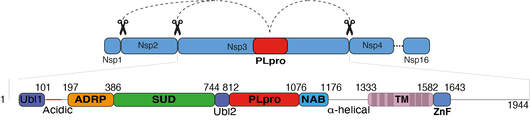
Armstrong L, Lange SM, et al. 2021 PLOS One
Of the 16 non-structural proteins (Nsps) encoded by SARS CoV-2, Nsp3 is the largest and plays important roles in the viral life cycle. Being a large, multidomain, transmembrane protein, Nsp3 has been the most challenging Nsp to characterize. Encoded within Nsp3 is the papain-like protease domain (PLpro) that cleaves not only the viral polypeptide but also K48-linked polyubiquitin and the ubiquitin-like modifier, ISG15, from host cell proteins. We here biochemically characterize Nsp3 and screen 1971 approved clinical compounds and identify five compounds that inhibit PLpro with IC50s in the low micromolar range but showed cross reactivity with other human deubiquitinases and had no significant antiviral activity in cellular SARS-CoV-2 infection assays. We therefore looked for alternative methods to block PLpro activity and engineered competitive nanobodies that bind to PLpro at the substrate binding site with nanomolar affinity thus inhibiting the enzyme. Our work highlights the importance of studying Nsp3 and provides tools and valuable insights to investigate Nsp3 biology during the viral infection cycle.
Final Full Text
Preprint
Of the 16 non-structural proteins (Nsps) encoded by SARS CoV-2, Nsp3 is the largest and plays important roles in the viral life cycle. Being a large, multidomain, transmembrane protein, Nsp3 has been the most challenging Nsp to characterize. Encoded within Nsp3 is the papain-like protease domain (PLpro) that cleaves not only the viral polypeptide but also K48-linked polyubiquitin and the ubiquitin-like modifier, ISG15, from host cell proteins. We here biochemically characterize Nsp3 and screen 1971 approved clinical compounds and identify five compounds that inhibit PLpro with IC50s in the low micromolar range but showed cross reactivity with other human deubiquitinases and had no significant antiviral activity in cellular SARS-CoV-2 infection assays. We therefore looked for alternative methods to block PLpro activity and engineered competitive nanobodies that bind to PLpro at the substrate binding site with nanomolar affinity thus inhibiting the enzyme. Our work highlights the importance of studying Nsp3 and provides tools and valuable insights to investigate Nsp3 biology during the viral infection cycle.
Final Full Text
Preprint

