Protein UFMylation
Ubiquitin fold modifier 1 (UFM1) is an 85-amino acid ubiquitin-like protein modifier (UBL) ubiquitously expressed in metazoans. UFM1 has low sequence homology to ubiquitin, but shares the characteristic beta-grasp fold of ubiquitin. Similar to ubiquitin and other UBLs, UFM1 is covalently attached to lysine residues of target proteins via a three-step enzymatic cascade catalysed by E1, E2 and E3 enzymes, UBA5, UFC1 and UFL1, respectively. The UFM1 pathway is linked to the production of membrane and secreted proteins in the endoplasmic reticulum and disruption of the UFMylation machinery triggers an ER stress response in cells. Membrane proteins perform essential functions and hence the biogenesis and quality control of membrane proteins at the ER are carefully controlled processes ensuring that abnormal and non-functional proteins which can be toxic to a cell do not accumulate. Hence, understanding how UFMylation regulates these pathways will advance our understanding of basic cell biology and reveal why failure in UFMylation results in ER stress and cell death. Indeed, homozygous disruption of the UFMylation pathway in mice causes embryonic lethality with defective hematopoiesis. Further, anomalies in the UFMylation pathway are linked to diverse pathologies including developmental defects, leukodystrophy, epileptic encephalopathy, spinocerebellar ataxia and skeletal abnormalities.
We therefore want to define how posttranslational modification of proteins with UFM1 maintains endoplasmic reticulum (ER) homeostasis by defining how UFM1 is conjugated onto distinct substrates in the ER by the E3 ligase, UFL1. Our main goals are to
1) Characterize at the molecular level how UFL1 functions as an enzyme to catalyse the transfer of UFM1 onto substrates, even though it has no recognizable ligase domains:
2) Understand how substrate specificity is achieved by adaptors that direct UFMylation onto specific substrates:
3) How is UFMylation modulated by deUFMylating enzymes?
4) How does UFMylation regulate protein translation and quality control at the ER?
We therefore want to define how posttranslational modification of proteins with UFM1 maintains endoplasmic reticulum (ER) homeostasis by defining how UFM1 is conjugated onto distinct substrates in the ER by the E3 ligase, UFL1. Our main goals are to
1) Characterize at the molecular level how UFL1 functions as an enzyme to catalyse the transfer of UFM1 onto substrates, even though it has no recognizable ligase domains:
2) Understand how substrate specificity is achieved by adaptors that direct UFMylation onto specific substrates:
3) How is UFMylation modulated by deUFMylating enzymes?
4) How does UFMylation regulate protein translation and quality control at the ER?
How are proteins UFMylated?
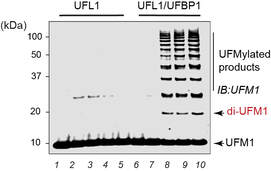
By using a rebuilding approach, we recently demonstrated that UFL1 together with UFBP1/DDRGK1 and CDK5RAP3 form a functional E3 ligase complex that specifically modify the ribosomal subunit RPL26 with UFM1. Our biochemical analyses reveal that this ligase complex is a scaffold-type enzyme that binds to the E2~UFM1 conjugate to activate aminolysis. Intriguingly, this E3 ligase complex bears no similarity to any of the known ubiquitin and Ubl E3 ligases. We are now trying to define the molecular mechanism of how this ligase complex functions to transfer UFM1 from the E2 specifically onto a single lysine residue on 60S ribosomes at the ER membrane.
Regulation of UFMylation
UFM1 is expressed as an 85 amino acid precursor that has to be post-translationally processed by the UFM1-specific thiol protease UFSP2 to remove two amino acids at the C-terminal end to generate mature UFM1 with a C-terminally exposed glycine. UFSP2 also performs an important regulatory role by cleaving UFM1 from modified proteins. Surprisingly, while UFM1 is an essential gene in several cell lines, cells lacking UFSP2 do not show any adverse effects. This is despite the fact that UFSP2 is required for producing functional mature UFM1. Hence it is likely that cells express other UFM1 specific proteases that have not yet been identified. Our recent work identifies the missing protease in human cells to be UFSP1 and we aim to understand the different roles of these two proteases in modulating UFMylation
